Model Calibration for Contamination Problem
MADS is applied to solve a general groundwater contamination problem using model inversion.
MADS includes analytical solver called Anasol.jl to solve the groundwater contamination transport in a aquifer.
Problem setup
import MadsSetup the working directory
cd(joinpath(Mads.dir, "examples", "contamination"))Load MADS input file
md = Mads.loadmadsfile("w01.mads")Dict{String, Any} with 7 entries:
"Grid" => Dict{Any, Any}("zmax"=>50, "time"=>50, "xcount"=>33, "zcoun…
"Sources" => Dict{Any, Any}[Dict("box"=>Dict{Any, Any}("dz"=>Dict{Any, A…
"Parameters" => OrderedCollections.OrderedDict{String, OrderedCollections.O…
"Wells" => OrderedCollections.OrderedDict{String, Any}("w1a"=>Dict{Any…
"Time" => Dict{Any, Any}("step"=>1, "start"=>1, "end"=>50)
"Observations" => OrderedCollections.OrderedDict{Any, Any}("w1a_1"=>OrderedCo…
"Filename" => "w01.mads"Plot
Generate a plot of the loaded problem showing the well locations and the location of the contaminant source:
Mads.plotmadsproblem(md, keyword="all_wells")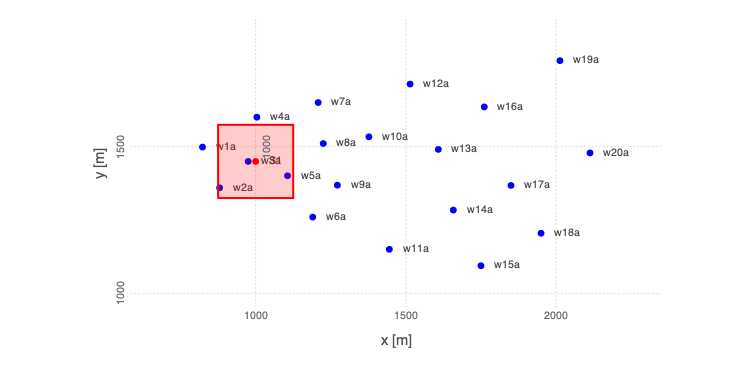
There are 20 monitoring wells.
Each well has 2 measurement ports:
- shallow (3 m below the water table labeled
a) and - deep (33 m below the water table labeled
b).
Contaminant concentrations are observed for 50 years at each well.
The contaminant transport is solved using the Anasol package in MADS.
Unknown model parameters
- Start time of contaminant release $t_0$
- End time of contaminant release $t_1$
- Advective pore velocity $v$
Reduced model setup
Analysis of the data from only 2 monitoring locations: w13a and w20a.
Mads.allwellsoff!(md) # turn off all wells
Mads.wellon!(md, "w13a") # use well w13a
Mads.wellon!(md, "w20a") # use well w20aOrderedCollections.OrderedDict{Any, Any} with 100 entries:
"w13a_1" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_2" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_3" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_4" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_5" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_6" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_7" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_8" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_9" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_10" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_11" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_12" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_13" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_14" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_15" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_16" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_17" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_18" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_19" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_20" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_21" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_22" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_23" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_24" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
"w13a_25" => OrderedCollections.OrderedDict{Any, Any}("well"=>"w13a", "time"=…
â‹® => â‹®Generate a plot of the updated problem showing the 2 well locations applied in the analyses as well as the location of the contaminant source:
Mads.plotmadsproblem(md; keyword="w13a_w20a")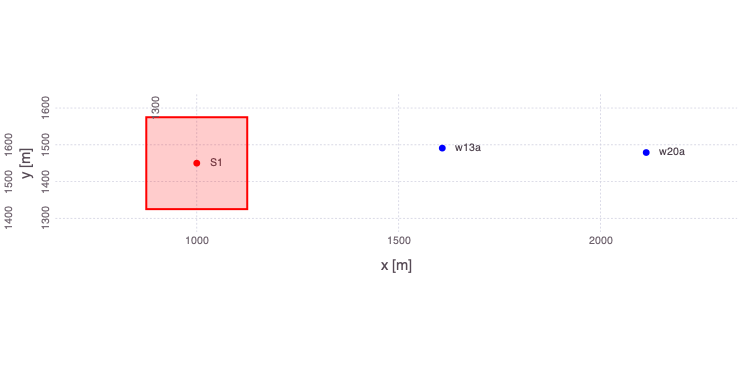
Initial estimates
Plot initial estimates of the contamiant concentrations at the 2 monitoring wells based on the initial model parameters:
w13a
Mads.plotmatches(md, "w13a"; display=true)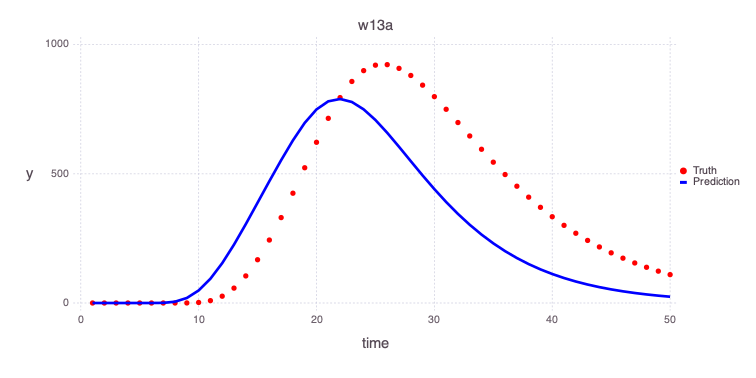
w20a
Mads.plotmatches(md, "w20a"; display=true)
Model calibration
Execute model calibration based on the concentrations observed in the two monitoring wells:
calib_param, calib_results = Mads.calibrate(md)Compute forward model predictions based on the calibrated model parameters:
calib_predictions = Mads.forward(md, calib_param)OrderedCollections.OrderedDict{Any, Float64} with 100 entries:
"w13a_1" => 0.0
"w13a_2" => 0.0
"w13a_3" => 0.0
"w13a_4" => 0.0
"w13a_5" => 0.0
"w13a_6" => 4.79956e-11
"w13a_7" => 0.000284228
"w13a_8" => 0.0590933
"w13a_9" => 0.92868
"w13a_10" => 5.08796
"w13a_11" => 16.2469
"w13a_12" => 37.7882
"w13a_13" => 71.6886
"w13a_14" => 118.275
"w13a_15" => 176.509
"w13a_16" => 244.465
"w13a_17" => 319.785
"w13a_18" => 400.036
"w13a_19" => 482.934
"w13a_20" => 566.28
"w13a_21" => 647.03
"w13a_22" => 720.732
"w13a_23" => 782.658
"w13a_24" => 829.249
"w13a_25" => 858.781
â‹® => â‹®Plot the predicted estimates of the contamiant concentrations at the 2 monitoring wells based on the estimated model parameters based on the performed model calibration:
w13a
Mads.plotmatches(md, calib_predictions, "w13a")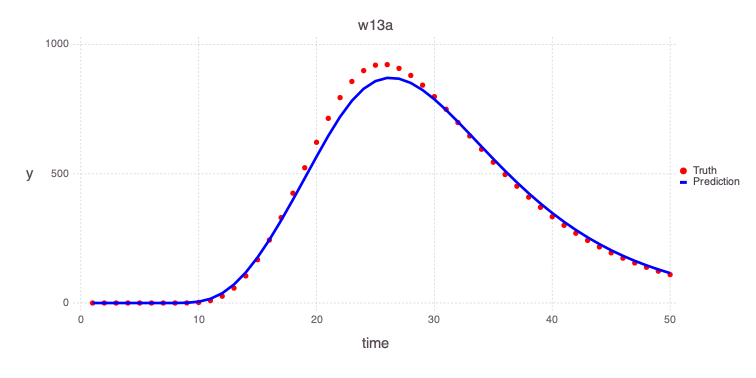
w20a
Mads.plotmatches(md, calib_predictions, "w20a")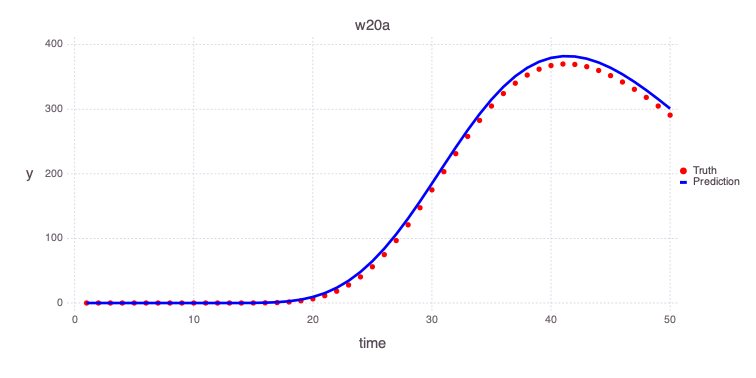
Initial values of the optimized model parameters are:
Mads.showparameters(md)Pore x velocity [L/T] : vx = 40 log-transformed min = 0.01 max = 200.0
Start Time [T] : source1_t0 = 4 min = 0.0 max = 10.0
End Time [T] : source1_t1 = 15 min = 5.0 max = 40.0
Number of optimizable parameters: 3Estimated values of the optimized model parameters are:
Mads.showparameterestimates(md, calib_param)3-element Vector{Pair{String, Float64}}:
"vx" => 31.059669248076222
"source1_t0" => 5.036614115598699
"source1_t1" => 16.62089724181972